pbso4-indexing
TOPAS-Academic – Tutorial
Peak fitting and Indexing of PbSO4
In this tutorial laboratory X-ray data of PbSO4 is indexed by first fitting peaks and then using those peak positions and intensities in an indexing run.
After selecting the data file, an input file for running topas with the same name but extension .inp is created in the same directory.
We will now use TA to get the initial peaks positions for peak fitting. From TA load the pbso4.raw file using the menu option “File/Import data files”. From inspection of the pattern is it seen that fitting to the x-axis region from 15 to 61 degrees 2Th should be sufficient for indexing.
From TA insert peaks using the menu option “View/Search peaks”. Inspect the inserted peaks and add/remove peaks such that their positions looks reasonable.
Expand the TA treeview to the “Peaks Phase” and click on the column heading called “Position”. Press the right mouse button on the yellow colored column and execute “Copy all/selection”. This copies the peak positions to the clip board. The TA screen should look like the following:
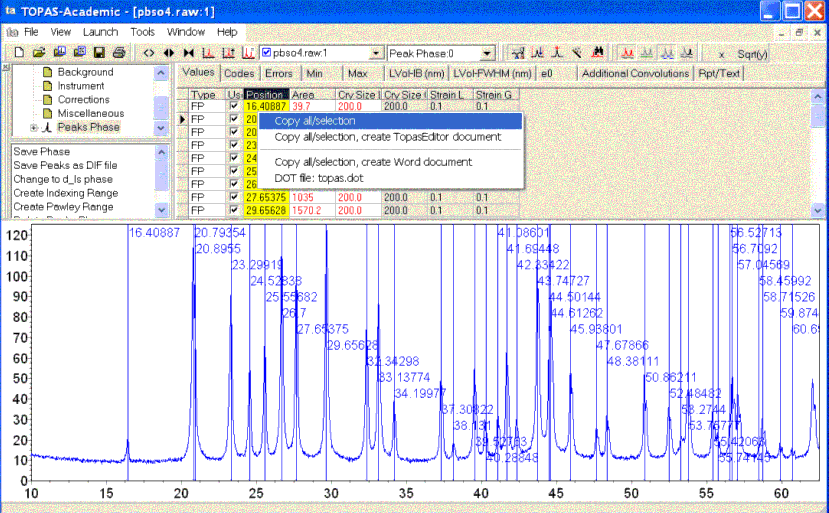
Paste the peak positions into jEdit.
Open the XInsert node “TOPAS-Academic, INP format/Scan file level”. Click on “Emission profile” and insert the CuKa5 emission profile.
Open the XInsret node “Background” and click on “Chubychev”.
Open the XInsret node “Instrument” and click on “Diffractometer Radius”, “Slit_Width”, “Divergence” and “Full_Axial_Model”; enter appropriate instrument values.
Open the XInsret node “Miscellaneous” and click on “start_X” and “finish_X” and enter the appropriate values.
Open the XInsret node “Phase level – xo_Is” and click on “xo_Is”, “CS_L”, “CS_G”, “Strain_L” and “Strain_G”.
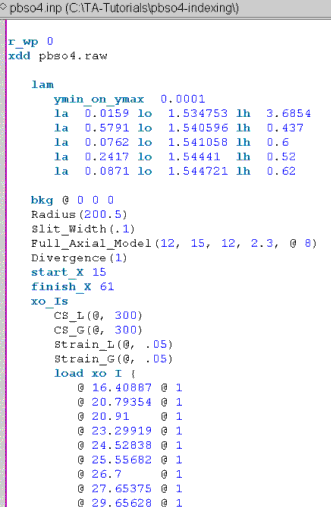
Enclose the peak psitions within braces in the text “load xo I { }”.
The jEdit screen should now look like the following:
Note, the User should typically know the instrument conditions corresponding to things like receiving slit width, axial divergence parameters, divergence, emission profile etc…
A zero error was not included as the peaks are free to move independently. A Lorentz Polarization factor was also not included as the peak intensities are not sought.
Also note that an xo_Is phase is used to fit the peaks and that the jEdit column cursor is used to insert the columns of ‘@’ and the intensities of ‘1’.
It may be useful at this stage to step through the jEdit column copying/cursor feature. If the INP keystrokes have been installed then to create a column cursor:
Press Ctrl-KeyPad_Plus; the jEdit status line should show an ‘R’ indicating that rectangular mode is on.
Hold down the Shift key and arrow down, you should see a column cursor.
Type the character @ and you should see a column of ‘@’ charatcers. Type another character and see what happens.
To cut a column press Ctrl-KeyPad_Plus, hold down the Shift key and use the arrow keys to select the desired rectangle. Press Shift-Del to cut the column. Move to where you would like the column pasted and press Shift-Ins.
To turn off rectangular mode then press Ctrl-KeyPad_Minus. Note, column copying/pasting works excellent in jEdit and it would be worthwhile to spend a minute or two becomming familiar with it. Under the XInsert node “TOPAS-Academic, INP format / Help” is “keyboard basics” which is useful to view.
Alt-Tab to TA and refine. The fit to the peaks should look like the following:
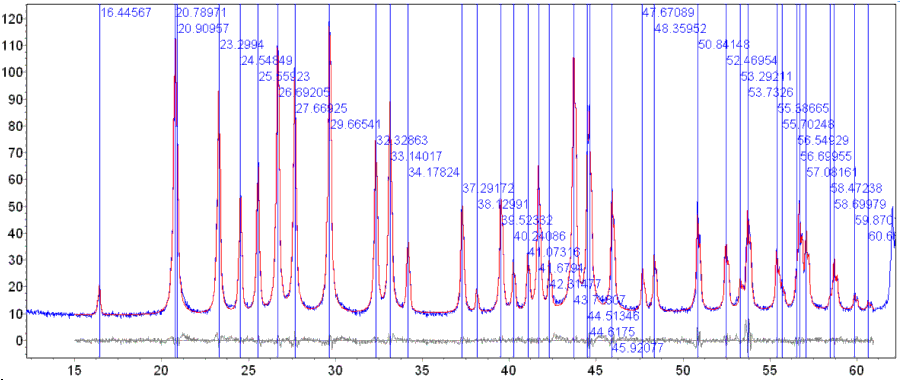
It is important to view the fit carefully and to make sure that peaks have not wandered off their approximate positions during the fit. The displayed vertical peak lines assists in this process. Note that the DEMO version of TA does not display the peak lines.
Indexing
We will now use the refined peak positions in an indexing run.
Save the pbso4.inp file to a file named indexing.inp. We will modify this file to perform indexing.
Delete everything from the INP file except for the peak positions and intensities.
Open the XInsert node called “Indexing” and work down the tree such that the INP file looks like the following:
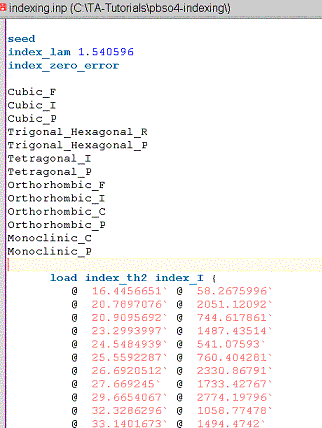
The indexing run produces a file with the same name as the INP file but with an NDX extension and in the same directory as the INP file. Load this file, INDEXING.NDX, into jEdit. The first part should look like the following:
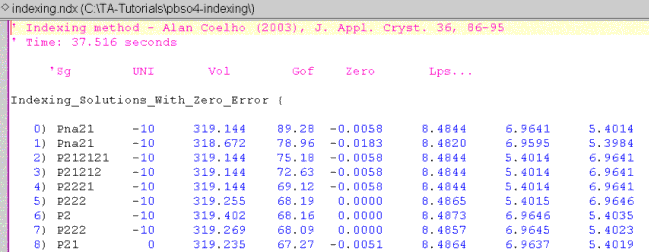
Here we see that the correct lattice parameters have been determined. Note that the space group for PbSO4 is Pbnm. The solution with the highest FOM is Pna21. Pna21 however has the same lines as Pbnm as far as powder data is concerned; see Table 12.6 of the Technical Reference. Thus the correct space group grouping was determined.
The solutions contained in indexing.ndx can be viewed graphically using TA.
First load indexing.inp into TA using the TA menu option “File/Import INP file”.
Copy the whole contents of the indexing.ndx file to the clipboard from jEdit. Alt-Tab to TA. Click on the Solutions Tab of the indexing range and execute the option “Paste INP to Node/Selections”. Click on one of the indexing solutions and the TA screen should now look like:
Note that high FOM values are observed for volumes that are multiples of the true solution of 319.
Note also that the loading of indexing.inp into TA would not be possible when keywords that are not understood by the GUI are used. In such cases then an indexing range can be manually created using the option “Create Indexing Range” found by clicking on the “Global” treeview item of TA. Once the indexing range is created then the 2Th/Intensity peak values can be copied from jEdit and pasted into the “Data” tab of TA. A window for displaying the indexing peak positions can be created using the TA menu option “Window/New Scan Window”. Thus graphical display of indexing solutions can always be easily displayed.
This completes peak fitting and indexing.
